Computational Structural Biology
Group Leader
| • Dr. Michael Habeck |
PhD Students
| • Nima Vakili |
| • Thach Nguyen |
Postdocs
| • Dr. Simeon Carstens |
Description
Many cellular processes are governed by macromolecular machines that are formed by large complexes of protein and nucleic acids. We develop computational methods to model and characterize these biomolecular structures at various levels of resolution. Using Bayesian statistics we integrate heterogeneous experimental data from NMR spectroscopy, cryo-electron microscopy, cross-linking / mass spectrometry, and chromosome conformation capture. To apply our approach in practice, we need efficient algorithms to sample all relevant conformational states of the system. Borrowing concepts and methods from statistical physics and machine learning, we have developed and implemented Markov chain Monte Carlo algorithms for probabilistic biomolecular modeling bundled in the Inferential Structure Determination (ISD) software. We have successfully applied ISD in a number of challenging structure determination projects such as the structure of a bacterial type 1 pilus. We recently started to use our methods for modeling the structures of chromosomes and genomes.Our research is funded by SFB 860 and part of the Felix-Bernstein Institute for Mathematical Statistics in the Biosciences (FBMS).
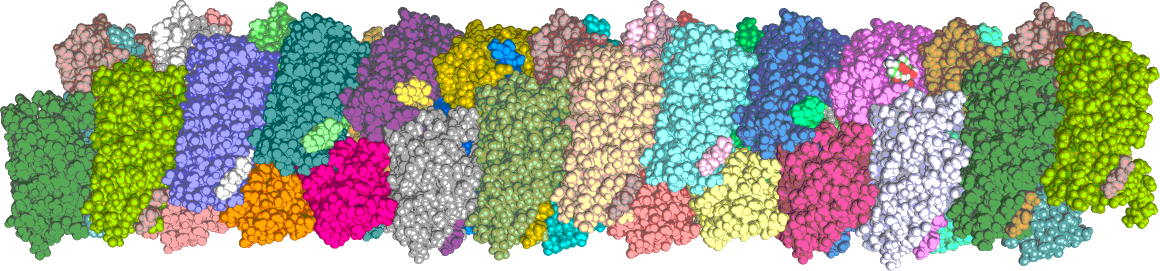
Near-atomic structural model of a bacterial type 1 pilus calculated with ISD.
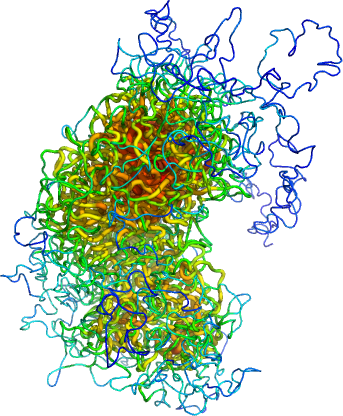
Model of the X chromosome obtained from single-cell HiC contact data.
Selected Publications
Data-driven coarse graining of large biomolecular structures.Chen YL, Habeck M
PloS One 2017; 12(8): e0183057
Inferential structure determination of chromosomes from single-cell Hi-C data.
Carstens S, Nilges M, Habeck M
PLoS Computational Biology 2016; 12(12): e1005292
Bayesian inference of initial models in cryo-electron microscopy using pseudo-atoms.
Joubert P, Habeck M
Biophysical Journal 2015; 108(5): 1165-1175
Hybrid structure of the type 1 pilus of uropathogenic Escherichia coli.
Habenstein B, Loquet A, ...,Habeck M, Lange A.
Angewandte Chemie 2015; 127(40): 11857-11861
Inferential structure determination.
Rieping W., Habeck M, Nilges M.
Science 2005; 309: 303-6

